Sequence Comparisons of 5S rRNA from P. aeruginosa, A. vinelandii and P. fluorescens to various tRNAs using a G:U Complement Intermediate
Alice C. Lichtenstein, M.S
aclsnippets@gmail.com.
Introduction
In a previous Snippets installment (www.DNARNAsnippets.com Snippets 11), the circular sequence of Peach Latent Mosaic Viroid (PLMVd) was compared, using a g:u intermediate (see methods below), to various known conserved sequences like reverse transcription tRNA primers, IgH V recombination signals, and origins of replication for mitochondrial and chloroplast DNA. The results were discussed in that particular installment, but, it was of interest to note, that one could find more than one "match" for a tRNA primer (approx 73 bp) along the whole PLMVd sequence as well as more than one match for the conserved sequences (CSB-1, CSB-2 and CSB-3) of the D loop of human mitochondrial DNA.
That particular result was either a total artifact derived from the empirical, subjective comparison or, a hint that there were underlying repeated sequences cached in the PLMVd sequence. The conserved sequences seemed to match best along the stems of double stranded RNA in the secondary structure of PLMVd, especially those of the human mitochondrial D loop; maybe, if the results are proven, a hint that "double-strandedness" was instrumental in conserving sequences.
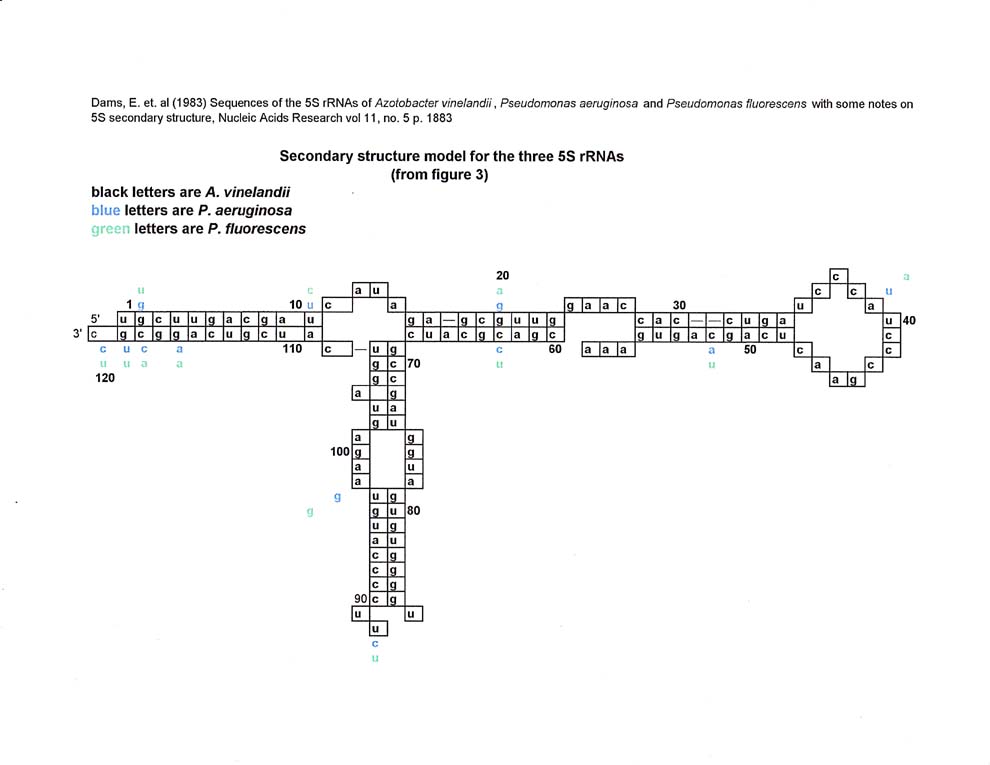
This author thought that if one used a smaller molecule to compare with tRNAs, the "matches" might be better. In looking for a suitable small sequence. it was decided to use 5S rRNA because it is found in most genomes at varying locations. The particular 5S rRNA sequence from A. vinelandii was chosen because the article (Dams et al (1983)) was readily available to download from the National Library of Medicine (www.ncbi.nlm.nih.gov)
Many articles show that tRNA molecules are created by splicing and ligating of "component parts" from larger pre-tRNA sequences. These articles describe two, three, and many other shorter sequences that are split out from chromosomes and subsequently joined together. One article describes the predicted formation of C. merolae tRNAs where the 3' part of the tRNA sequence is reported to be upstream of the 5' part and a circular intermediate is needed to produce the correct 5' to 3' final sequence. (Soma, A et. al. (2007)).
Another article demonstrates that some pre-tRNAs contain introns that are homologous to sequences known to be split out from different DNA locations and then ligated together to form the completed tRNA. (Fujishina, K. et.al (2009)).
Mark, C and Grosjean, H. (2003) show 18 archaea pre tRNAs, whose most common intron splice site is at 37/38 (at the anticodon). However, they also have intron splice sites at 14 other base junctions (3/4, 20b/21, 21/22, 22/23, 28/29, 29/30, 30/31, 32/33, 38/39, 39/40, 45/46, 56/57, 58/59, 59/60).
Thus it is clear that there are many ways to generate tRNAs from pre-tRNAs. Keeping that in mind, the comparisons were done as follows.
Methodology
The sequences of 5S rRNA from Pseudomonas aeruginosa, Azotobacter vinelandii and Pseudomonas fluorescens were compared to various tRNA sequences from homo sapiens, chloroplasts, mitochondria, archaea and circular C. merolae, using a g:u complement. Later, a comparison of 5SrRNA was also done to recombination signals from T cell receptor beta chain and recombination signals used in VDJ Immunoglobulin formation.
As described in previous installments of Snippets a g:u complement was created as follows :bases g and u were used to make a complement to a sequence then a reverse complement to the gu complement was made. In other words, purines could be substituted for purines and pyrimidines for pyrimidines in the original sequence to see if there was a "match" between 5S rRNA and the specific tRNA being compared to it.
| sequence | C | C | A | G | G | G |
| gu complement | G | G | U | U | U | U |
| reverse complement | U or C | U or C | A or G | A or G | A or G | A or G |
As an example, the following matches were derived by comparing the first eight bases of 5S rRNA from P. aeruginosa to tRNA lys 3 ( a reverse transcriptase primer), tRNA met (tobacco chloroplast), tRNA leucine (homo sapiens mitochondria), tRNA glycine (archaea C. maquilensis).:
| 5S rRNA (P.a.) | U | G | C | U | U | G | A | #1-7 |
| g:u complement | G | U | G | G | G | U | U | |
| tRNA lysine3 | G | C | C | C | G | G | #1-6 | |
| tRNA fmet (tobacco chloroplast) | G | U | U | U | G | A | #18-13 | |
| tRNA leucine (mitoch.) | G | C | C | C | G | A | #19-14 | |
| tRNA glycine archaeum | C | G | C | C | G | G | #72-66 |
More than one match was made for some of the tRNAs,
The tRNAs were chosen randomly because the author wanted to see a variety of comparisons, and if there was any consistency, it would be followed up.
The sequence for 5S rRNA that was used for comparisons is below:

Comparisons of the complete tRNA sequences are reported below. Please note that the g:u complement for 5S rRNA is not displayed in the figures.
Results
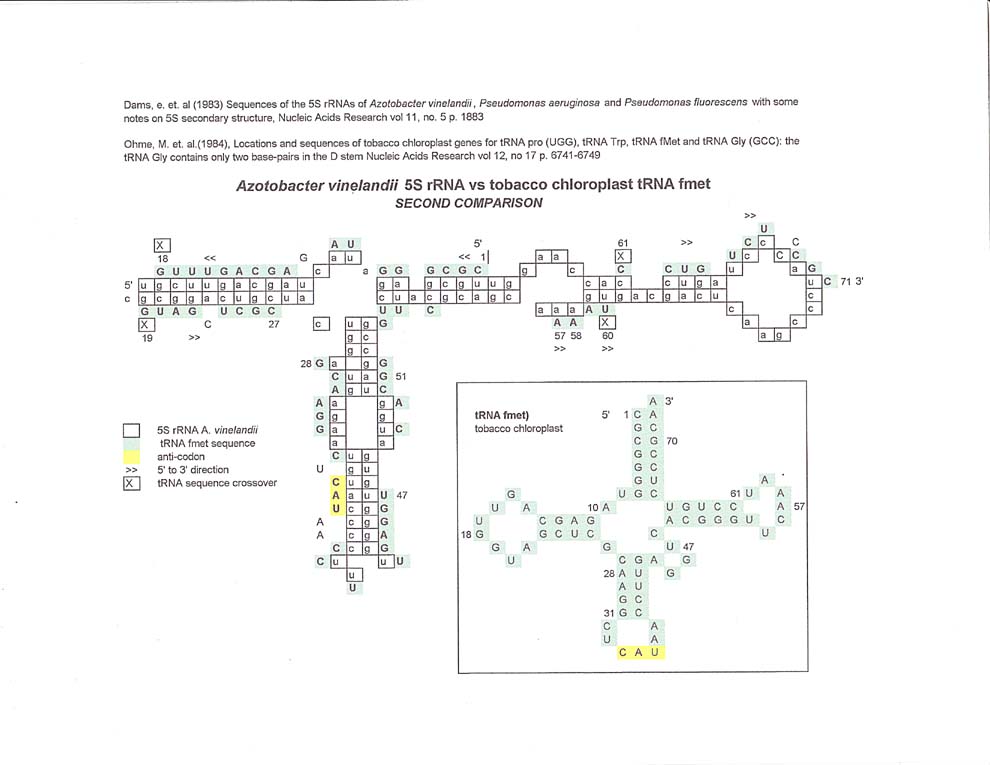
5S rRNA vs tobacco chloroplast tRNAs
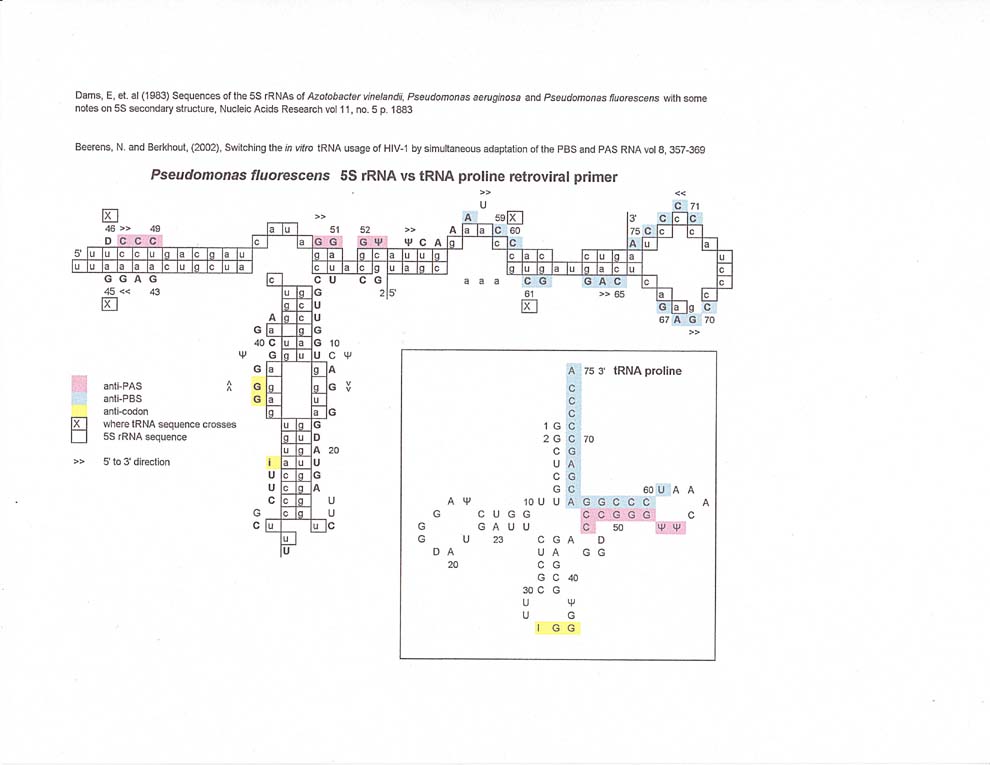
5S rRNA vs retroviral primer tRNAs
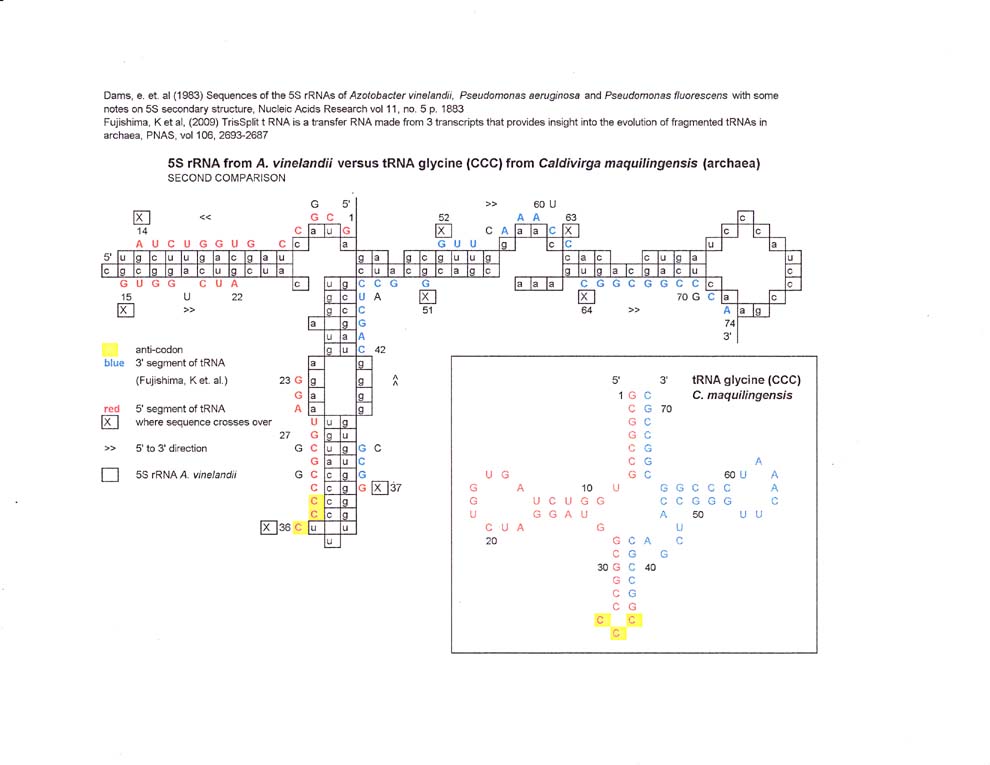
5S rRNA vs. C. maquilingensis tRNA
5S rRNA vs tobacco chloroplast tRNAs
The matches do not seem to be random, even though there is a 50:50 chance at each rRNA base that there will be a match with a tRNA base. Rather, the aligned sequences seem to be in segments of 4 or more bases with discrete gaps in between. The gaps, where there are no matches will be referred to as "spacers" for the sake of discussion.
Thus, looking at the tobacco chloroplast tRNAproline alignment below, bases 1 thru 9 are aligned with one mismatch, 10-16 are aligned with one mismatch, 17 through 26 are aligned with one mismatch, 27 thru 35 are aligned with one mismatch, and 36 thru 51 are aligned with 2 mismatches. Then there is a gap in the alignment of 14 bases followed by 52 thru 60, all matches, a gap of 5 pyrimidines and then 61 thru 72 with 4 mismatches. This author cannot say anything about the fact that the 3' end of the tobacco chloroplast tRNA proline sequence abuts the 5' end of the sequence other than the sequence alignment follows a somewhat circular pattern around the 5S rRNA sequence.
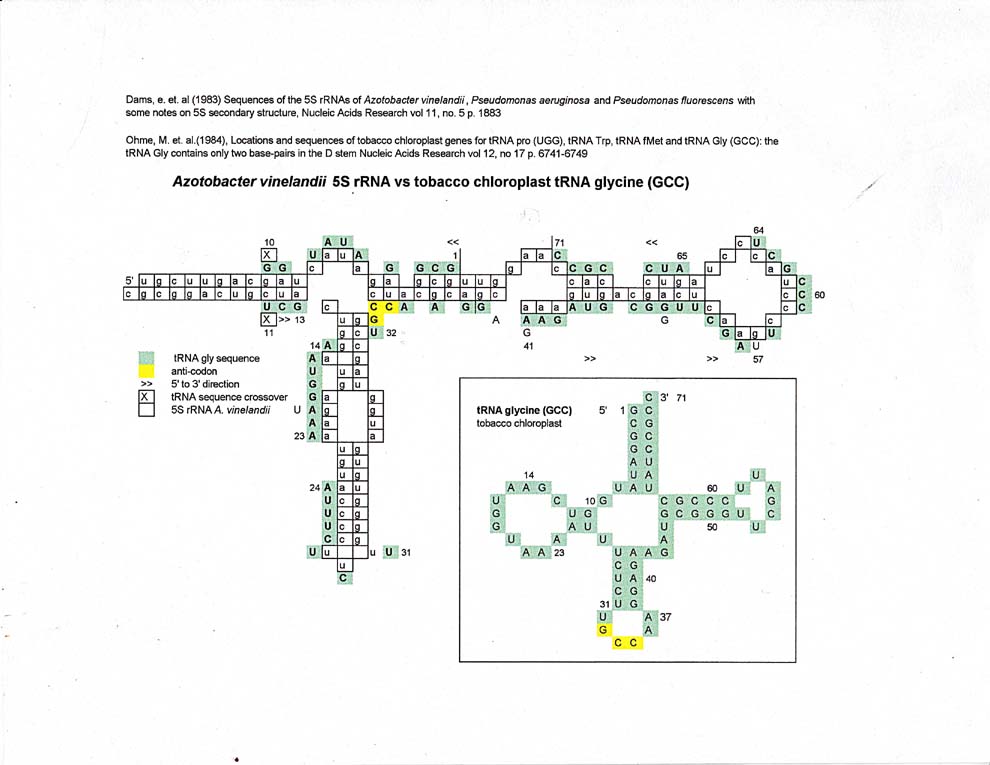
The tobacco chloroplast tRNA glycine alignment with 5S rRNA has more mismatches, but once again, one can see alignments of discrete sequences in a circular pattern around the 5S rRNA sequence.
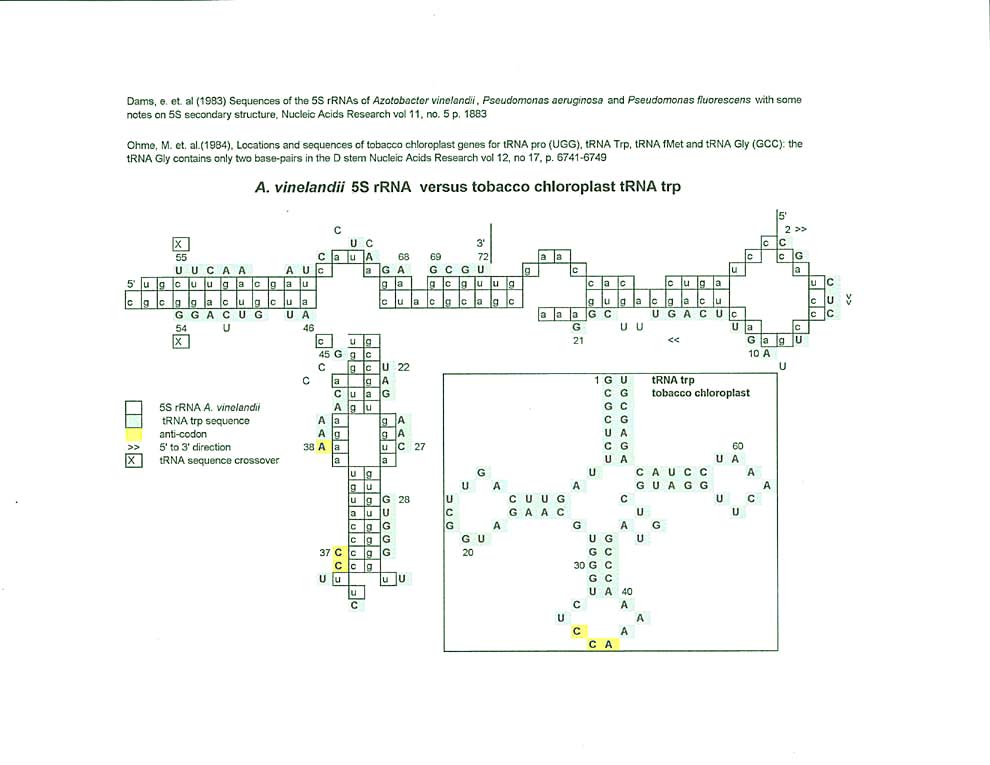
Tobacco chloroplast tRNA trp can be aligned with 5S rRNA going in the opposite direction from the other two tobacco chloroplast comparisons. 2 thru 21 has 3 mismatches, then there is a 13 base gap between 21 and 22, between 22 and 37 there are 4 mismatches, then a 7 base gap.etc. Even though there are a number of mismatches between 38 and 54 the alignment can still be determined to follow a circular pattern and ends with a gap between the 3' and 5' ends of the tRNA.
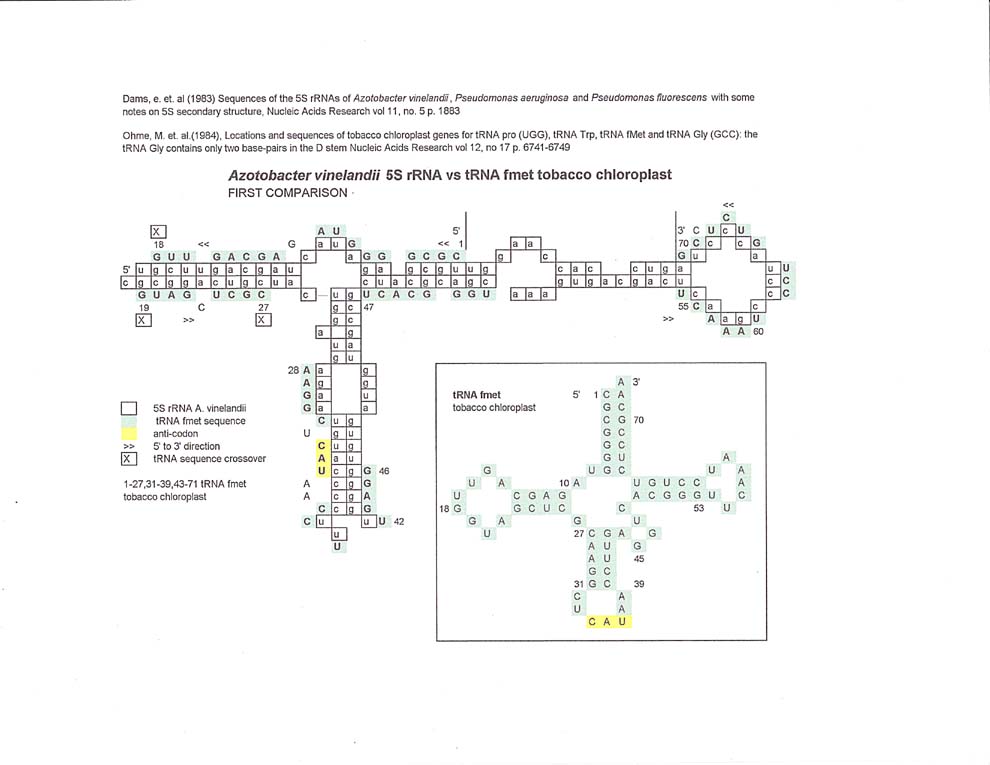
The tobacco chloroplast tRNA fmet comparisons show that there can be more than one alignment between the two molecules. The first comparison follows a circular pattern, but the second alignment shows a sequence crossover between tRNA bases 60 and 61. Of course this is due to the subjective methodology in making the alignments, but might also reflect some sort of underlying sequences in the rRNA molecule.
As will be seen below, the tobacco chloroplast tRNAs have the best alignments of all the tRNAs compared to the 5S rRNA sequence.

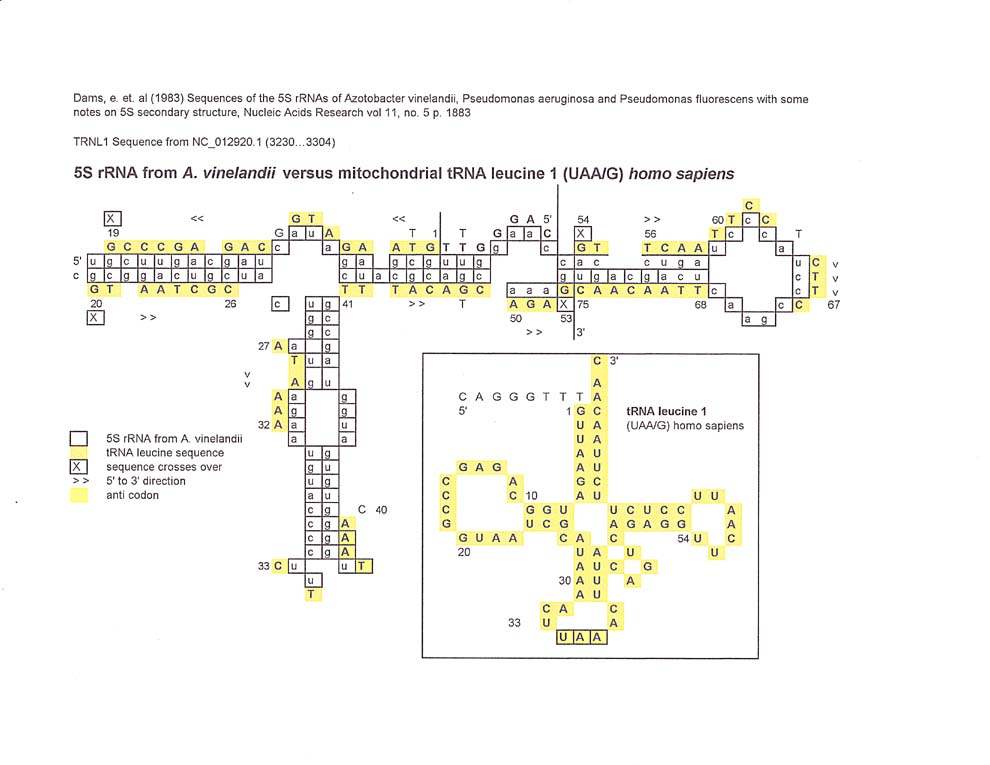
Of interest in the comparison between A. vinelandii 5S rRNA and mitochondrial tRNA leucine 1, is the match along the eight bases 5' to the start of the tRNA sequence. There is one crossover of bases between 53 and 54.
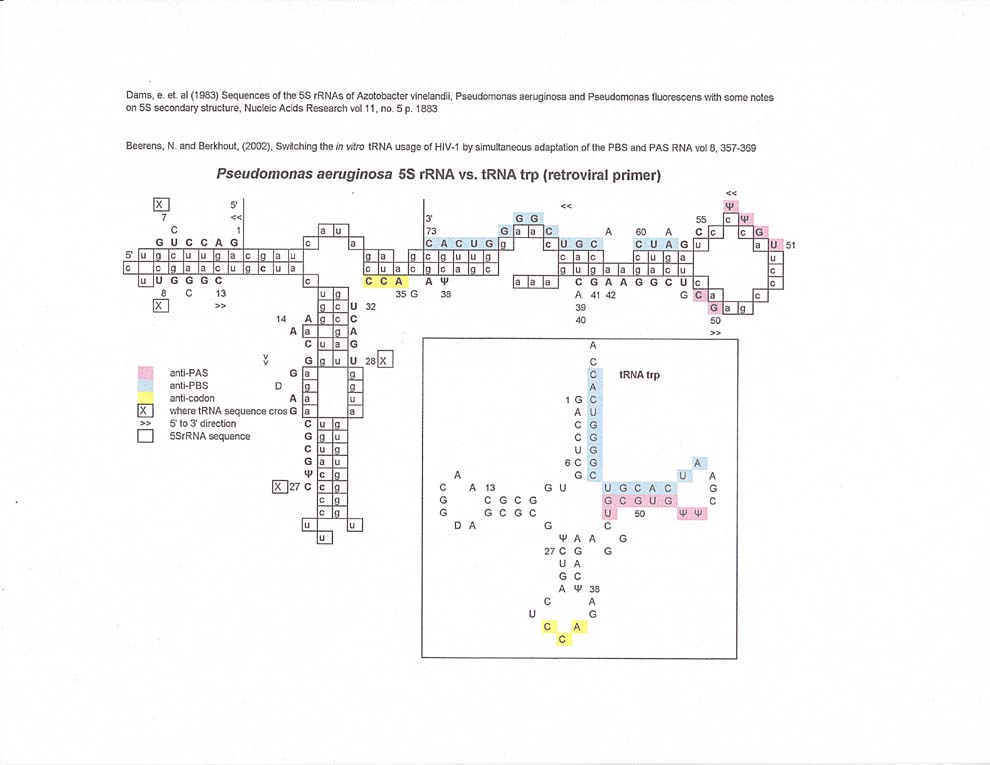
5S rRNA vs retroviral primer tRNAs
The match along 5S rRNA for tRNA trp (retroviral primer) is somewhat circular with both the antiPAS and anti PBS sequences on one arm of the 5S rRNA. In fact, the tRNA proline and tRNA lysine3 matches for anti PBS and anti PAS also extend along the same arm.
(As an aside, the fact that tRNA trp's bases #1-7 are complementary to #8 through 13 may hint at a switch function in the tRNA molecule.)

tRNA ala(UGC) yielded two results. The matches from the first comparison once again seem to be in discrete sequences with gaps in between. 1 through 12 are a perfect match and 13 through 26 have 3 mismatches. The crossover at 26 to 27 extends to another crossover from 33 to 34. After that the matches aren't as good as seen previously; 34 to 42 has 2 mis matches, 43 to 73 has 10 mismatches.
Also, the ligation site predicted in Soma et al.'s article (where the bases indicated in blue are ligated to the bases indicated in red) does not coincide with any discrete gap in the 5S rRNA alignment. That was to be expected.
However, tRNA bases 1 through 12 and 13 through 26 that are mostly continual along the "vertical" arm (for want of a better description) of the rRNA sequence, correspond to the 5' sequence of the acceptor arm as well as the D arm of the tRNA. 27 through 42 correspond to the anticodon arm, and following a gap, 43 through 69 correspond to the extra arm, the TΨC arm and part of the 3' end of the acceptor arm.
This probably does not mean anything in terms of a biological mechanism.
%20vs%205S%20rRNA%20first%20comparison%2030%20percent.jpg)
%20vs%205S%20rRNA%20second%20comparison%2030%20percent.jpg)
5S rRNA vs. C. maquilingensis tRNA
In the first comparison of C. maquilingensis tRNA glycine vs
5S rRNA bases 1 thru 8 have one mismatch and then the sequence crosses over and
continues. At the same place, bases 72 to 66 cross over too and the crossovers
correspond to the acceptor stem of the tRNA. Otherwise the sequence matches
follow a "circular" pattern around the 5S rRNA.

Discussion
The problem with the comparisons is that one cannot deduce anything from them in terms of biological significance. They don't answer the question "Is there any relationship between 5S rRNA from A vinelandii and the various tRNA sequences used?" Or, were the 5S rRNA and tRNA sequences originally circular?" Or, does the g:u complement used to generate the matches reflect anything real? It is very important to note, however, that using the g:u complement does generate alignments.
Since, for any given comparison, there was usually one or more discrete "spacers" along the alignments it was decided to look into these further to see if they corresponded to any known splice sites or known pre-tRNA intron sequences. The "spacers" from tobacco chloroplast tRNA comparisons were used since, subjectively, their alignments appeared to be the "best fit" to 5S rRNA. (Author's note: if there were matches to other known conserved sequences, the author included those too.)
The results below show a comparison of the individual tobacco chloroplast tRNA "spacers" with reported splice sites or introns from circularized pre-tRNAs. The C. merolae sequences were from Soma, A et. al. (2007), the P. aerophilum and T. neutrophilus sequences were from Fujishina, K. et.al (2009).
Some of the results were intriguing as shown in the figure(s) below.
Tobacco chloroplast tRNA glycine, tRNA fMet1 and tRNA trp all had a "spacer" between their first base (no. 1) and their last base along the same region of the rRNA sequence. The "spacer" was reminiscent of the sequence between the 3' and 5' circular form in C merolae (Soma, et al. 2007) so a comparison was done. In the result below, the tobacco chloroplast tRNA "spacers" are shown at the top of the appropriate rRNA sequence region, and the known tRNA splice sites and introns are below.
The 5' start site of tobacco chloroplast tRNA glycine (CCC) and tobacco chloroplast tRNA Met1, and the 3' end of tobacco chloroplast tRNA trp, C. merolae tRNA threonine (AGT), C. merolae tRNA Gln (CTG) and C. merolae tRNA arginine (CCT) align (to within one base) to base number 21 of 5S rRNA. The splice site at base number 29 of tRNA fMet P. aerophilum and tRNA threonine of T. neutrophilus also align at base 21 of 5S rRNA.
The match between the loop that is clipped out of C. merolae's tRNA threonine and the spacer between tobacco chloroplast tRNA trp seem to line up fairly well. But, again, one cannot tell if the tobacco chloroplast "spacer" really exists or existed or is an artifact of the method used to compare sequences.
Also, that while the region between numbers 18 and 49 align with "spacers" for these particular chloroplast tRNAs, for other tRNAs it aligns with the actual tRNA sequence. Note especially retroviral primer tRNAtrp where the antiPBS region aligns with numbers 19 to 37. In other words, the matches can be both along a "spacer" or part of a tRNA.

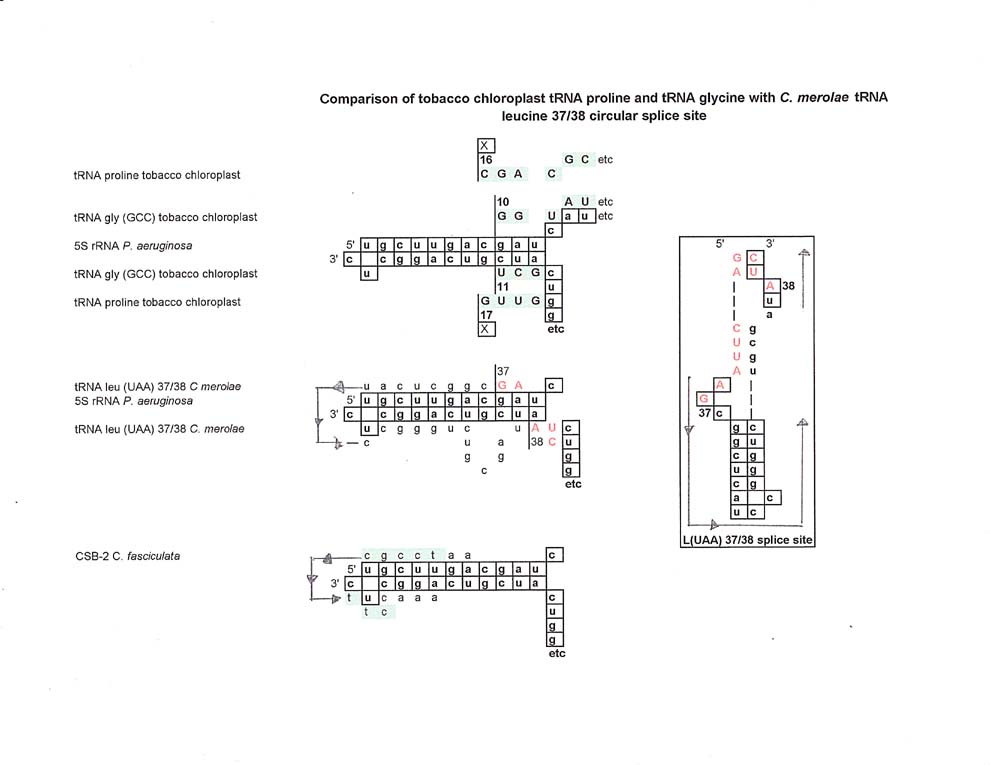
In a comparison of tobacco chloroplast tRNA proline and tRNA glycine with C. merolae's tRNA leucine 37/38 splice site, similarities are found, especially if one visualizes that the 5' and 3' ends of the 5SrRNA sequence are ligated, generating a "circular" version of 5S rRNA. Shown below are the 5' and 3' ends of 5S rRNA and the alignments where the sequences of tRNA proline and tRNA glycine "cross over" the 5S rRNA strands. As shown, there are similarities in location and sequence to the C. merolae tRNA leucine 37/38 loop splice site.
For the sake of comparison there are two more non-tRNA sequences that are aligned: CSB-2 from Crithidia fasciculata
CSB-2 is conserved, with some minor variation in the mitochondrial minicircles of most kinetoplasts. It's function is not really known.
Comparisons of 5S rRNA and known pre-tRNA introns with TCR recombination signals
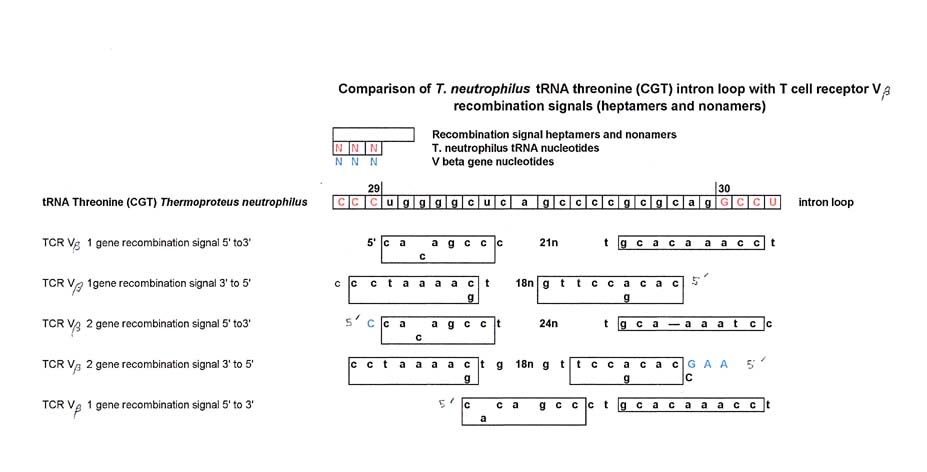
At this point we have to make a digression and talk about another sequence that aligns with Thermoproteus neutrophilus tRNA threonine's intron loop: the conserved heptamer (octamer?) and nonomer sequences that are included in the recombination signals of some T cell receptor (TCR) V beta chain genes. (see two figures above for T neutrophilus tRNA threonine intron loop). By implication, they also align with 5S rRNA base number 21 through about base number 40.
For our purposes the recombination signal is used to clip out a specific V gene that is to be used in the assembly of a completed TCR beta chain. The 5' end of the heptamer follows the 3' end of the particular V gene and is involved with splicing the V gene out of its place in the DNA. It is not unlike the mechanism for VDJ recombination in immunoglobulins. (Krebs, J. et. al. (2011) Lewin's Genes X)
In the figure below, the intron region that is spliced out of pre-tRNA threonine (CGT) from T. neutrophilus at 29/30, (that was shown above in a comparison with 5S rRNA) is now shown in a comparison with the recombination signals for TCR V beta 1 and V beta 2 genes. The V beta gene recombination signal sequences are from Rowen, et. al (1996). The 5' end of the heptamer lines up with the 29 base splice site of pre-tRNA threonine (CGT). , but the alignment is not as good for the 30 splice site.
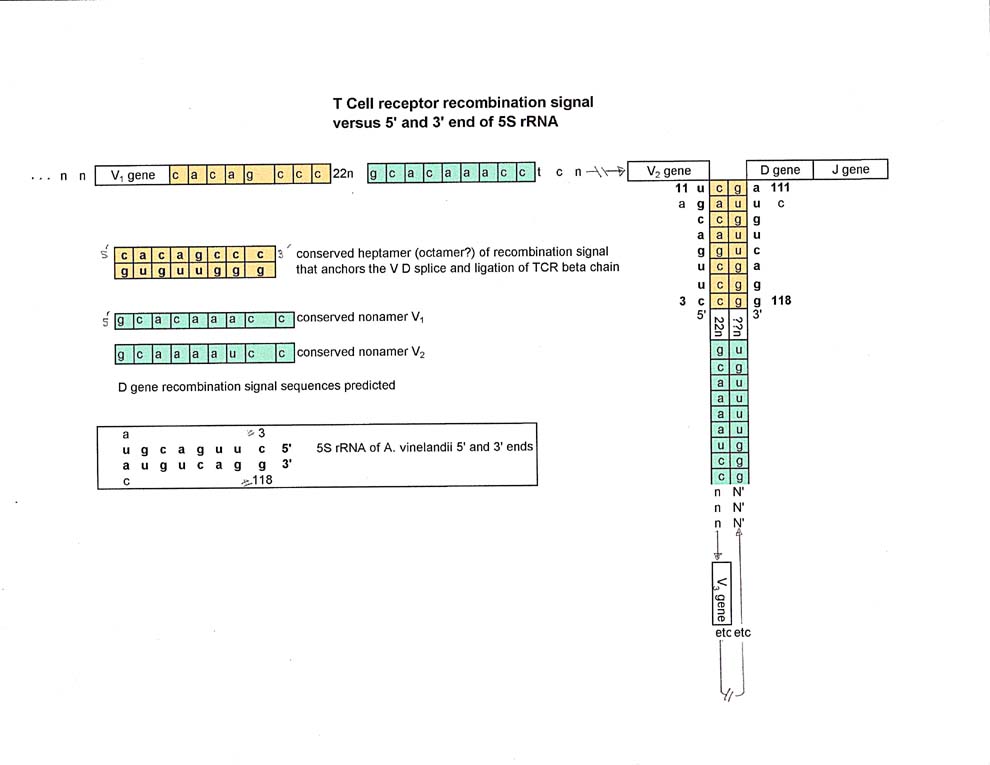
More startling was the recognition that the "spacer" for tRNA glycine and tRNA proline that aligned with the 5' and 3' ends of the 5S rRNA also very closely aligned with the heptamer (octamer?) from the TCR V beta chain recombiantion signal.
The TCR V genes that are used in the formation of, in this case, beta chains are all followed at their 3' end by a recombination sequence that starts with CACAGYYY Although Rowen et. al. posited that the sequence was really CACAGTG like the recombination signal for Immunoglobulins, this author, after looking at all the sequences published by Rowen et al , used the octamer CACAGYYY and mostly CACAGCCC. Below is a comparison of the octamer and the 5S rRNA 5' and 3' ends.
The tRNA ala (TGC) C. merolae splice site at base 59 is (3')...caUAGCUU... (5')
lower case letters are the intron of the predicted circular sequence of pre-tRNA ala (TGC)
Upper case blue letters are the exon sequence
| tRNA ala TGC C.m. | c | a | U | A | G | C | U | U |
| g:u complement | g | u | g | u | u | g | g | g |
| V 4-1,2,3 | c | a | c | a | g | c | c | t |
| V 5-1, 3 | c | a | c | a | g | c | c | c |
| V 8-1,2 | c | g | c | a | g | c | c | c |
| V 9 | c | a | c | a | g | c | c | c |
Other cleavage site comparisons produced interesting results but none involved loops that matched "spacers" from chloroplast tRNAs.
Below is a comparison of the "spacer" between numbers 31 and 32 of tobacco chloroplast tRNA glycine and that of pre-tRNA glycines from C. merolae and C. maquilingensis and pre-tRNA P. islandicum. The 37 and 38 splice sites of tRNA glycine of C. merolae and C maquilingensis line up to within one base of the 31 "spacer" site, but this doesn't mean anything for now. All that can be said is that there are similar alignments of sequences among the various tRNA glycines and the region of 5S rRNA when seen through a gu complement. It would be nice to find a pre-tRNA sequence for tobacco chloroplast tRNA glycine in the literature.
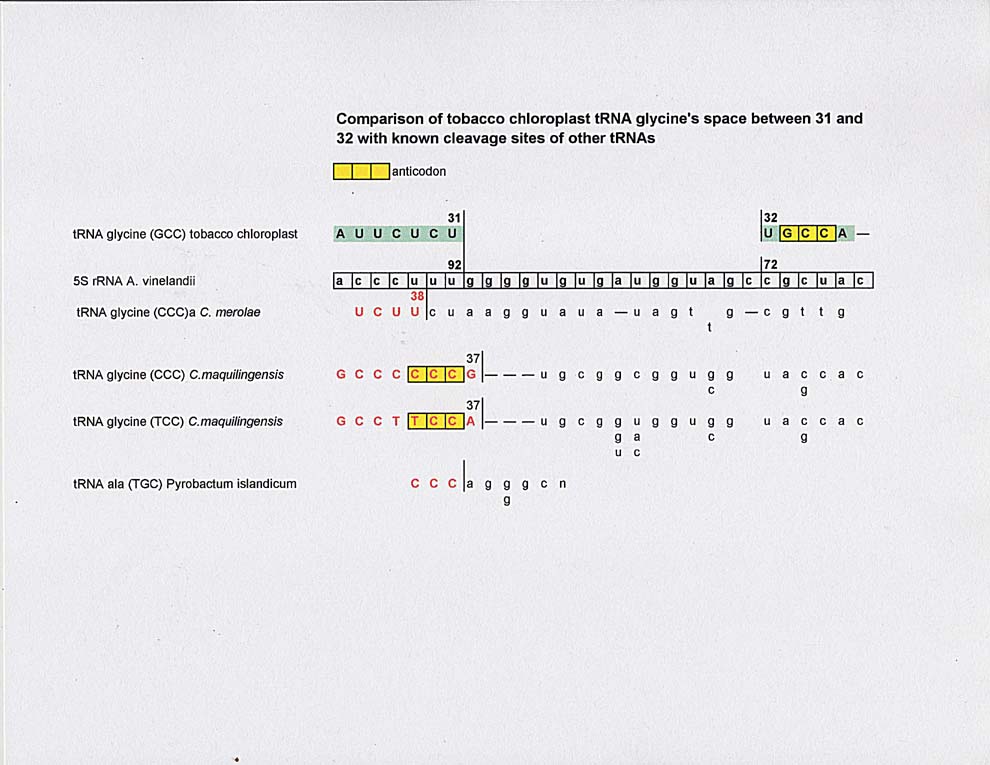
So, what does this all mean, if anything? It is hard to tell.
The g:u intermediate is a device this author used as a way of finding other sequences but does not mean a "g:u" alphabet actually existed at any time.
Are there sequences that use both sets of pyrimidines or purines? The only set of sequences this author could find in the literature available online, were gRNAs in kinetoplasts. The gRNAs of kinetoplast mitochondria, are generated from a set of minicircles, and are used as a template to insert U bases in mRNA transcripts from kinetoplast maxicircles. For our purposes, this author will not go into the mechanism.
According to the figures in Gao, G et.al (2001) the gRNAs are lined up with overlapping ends 5', along the maxicircle. Many of these gRNAs are redundant. Where the gRNAs overlap in sequence, one can see that both pyrimidines, or both purines are used as a template for U insertion. (see below from figure 2) The table below shows where gND9-V and gND9-III overlap gND9-IV and show different purines and pyrimidines. (Bolded letters show a different purine or pyrimide base.) The overlaps between gND9-IV, -V and -III are homologies, not complements.
|
from figure 2 ..."overlapping gRNAs mediating the editing of the gND9 mRNA in the LEM 125 strain..." (Gao, G. et. al. (2001)) |
|||||||||||||||||||||||||||||
| gND9-IV | ETC | G | A | U | A | U | A | U | C | A | A | G | C | A | 12n | A | C | A | U | A | C | A | A | U | A | C | A | 5' | |
| gND9-V overlap | ETC | A | A | C | A | (a) | A | C | C | A | A | A | C | A | 5' | ||||||||||||||
| gND9-III overlap | G | C | A | U | A | U | G | A | U | A | U | G | ETC | 5' | |||||||||||||||
| maxicircle U insertion | 5' ETC | u | u | g | u | u | u | g | g | u | u | u | g | u | nn... | u | g | u | a | u | g | u | u | a | u | g | u | ETC | |
ETC means that the sequence continues in the 3' or 5' direction.
For what it's worth, as a complete aside, the overlap region of gND9-III, using a G:U intermediate is CACAGTG the heptamer recombination signal for immunoglobulins, and the overlap region of gND9-V has a truncated sequence similar to the nonamer recombination sequences of both immunoglobulins and TCR. IgH sequences are from Kefei, Yu et. al. (2002)
|
from figure 2 ..."overlapping gRNAs mediating the editing of the gND9 mRNA in the LEM 125 strain..." (Gao, G. et. al. (2001)) |
|||||||||||||||||||||||||||
| gND9-IV | ETC | G | A | U | A | U | A | U | C | A | A | G | C | A | 12N | A | C | A | U | A | C | A | A | U | A | C | A |
| G:U Complement | U | U | G | U | G | U | G | G | U | U | U | G | U | U | G | U | G | U | G | U | U | G | U | G | U | ||
| IgH Heptamer | G | T | G | A | C | A | C | 5' | |||||||||||||||||||
| IgH Heptamer | 5' | C | A | C | A | G | T | G | |||||||||||||||||||
| IgH Nonamer(part) | 5' | C | C | A | A | A | C | A | |||||||||||||||||||
This is different than the RNA editing described by Yoshinaga, K et. al (1996) where genes from chloroplast germline DNA were compared to cDNA of their mRNA transcripts and U to C as well as C to U substitutions were noted. These changes were made post-transcriptionally.
The author understands that using the G:U intermediate method one can find many sequences derived from one original sequence. Whether this is an artifact of exchanging pyrimidines and exchanging purines or reflects some evolutionary process remains to be seen.
References
Beerens, N and Berkhout, B (2002), Switching the in vitro tRNA usage of HIV-1 by simultaneous adaptation of the PBS and PAS, RNA, vol 8, pps. .357-369
Dams, Erna et al, (1983), Sequences of the 5S rRNAs of Azotobacter vinelandii, Pseudomonas aeruginosa and Pseudomonas fluorescens with some notes on 5S RNA Secondary Structure, Nucleic Acids Research, vol 11 No 5, pps 1245-1252
Fujishima, K et. al, (2009), Tri-Split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea, PNAS vol 106, pps 2693-2687
Gao, G. et. al. (2001), Guide RNAs of the recently isolated LEM 125 strain of Leishmania tarentolare: an unexpected complexity, RNA vol 7, pp 1335-1347.
Kefei, Yu et al (2002), The Cleavage Efficiency of the Human Immunoglobulin Heavy Chain VH Elements by the RAG complex, JBC vol 277, pp5040-5046.
Krebs, J. et. al, (2011) Lewin's Genes X, Jones and Bartlett Publishers, LLC
Mark, C and Grosjean, H (2003) Identification of BHB Splicing motifs in intron-containing tRNAs from archaea: evolutionary implications, RNA vol 9, 1516-1531
Ohme, M. et al, (1984), Locations and sequences of tobacco chloroplast genes for tRNA Pro (UGG), tRNA Trp, tRNA fMet and tRNA Gly (GCC): the tRNA Gly contains only two base-pairs in the D stem, Nucleic Acids Research, vol 12, no 17, pps. 6741-6749
Ray, D (1989), Conserved Sequence Blocks in Kinetoplast Minicircles from Diverse Species of Trypanosomes, Molecular and Cellular Biology vol 9, no 3 pps 1365-1367.
Rowen, et. al. (1996), The Complete 685 kilobase DNA sequence of the Human beta T cell receptor locus, Science Vol 272 p. 1757
Soma, A., et. al. (2007) Permuted tRNA Genes Expressed via a Circular RNA Intermediate in Cyanidioschyzon merolae, Science vol 318, pps. 450-453, errata 30 November 2007 and 21 Dec 2007.
Yoshinaga, K. et. al. (1996) Extensive RNA editing of U to C in addition to C to U substitution in the rebcL transcripts of hornwort chloroplasts and the origin of RNA editing in green plants, Nucleic Acids Research, vol 24, pp1008-1014.