
On the Sequence Similarity of some Illegitimate Translocation Breakpoints and the RNA -Primed Origins of Replication for H and L strands of Mitochondrial DNA Kinetoplast Minicircles
Alice Lichtenstein, M.S.
aclichtenstein@nyc.rr.com
This is a continuation of Snippets9a.html where the results of comparisons between bcl6 and bcl translocations and nucleotide sequences of origins of replication of kinetoplastid DNA minicircles were reported.
Discussion
General Comments
If two compared sequences are similar, it does not mean necessarily that one sequence is derived from the other; therefore, one cannot say that kinetoplastid mitochondrial minicircle DNA gave rise to these somatic translocations without further comparisons and direct scientific experimental proof. Most probably, these sequences occur in other molecules too, and a search for these sequences in tRNAs, and archael pretRNAS revealed the following results
In the paper by Abu-Eineel et. al., that characterized the Universal Minicircle Sequence-binding Protein, it was mentioned that minicircle H strand and L strand initiation sites were between 70-100 nucleotides apart. That, and the fact that the number of nucleotides that separates CSB-1, 2 and CSB-3 is roughly 100 encouraged this author to look at the sequences of pre-tRNAs and tRNAs more systematically and search for CSB and oril and oriH-like sequences. If one was willing to use RNA complements that allowed for g:u bonds, a pattern emerged in the following examples.
Marck and Grosjean looked closely at intron/exon sites in archael tRNAs and found that besides the previously noted 37/38 anti-codon sites, characterized by a bulge-helix-bulge, there were others. Without going into details of their findings, the pre-tRNAs shown below , from their figure #3, were visually scanned for Ori H and Ori l sequences. Complementary sequences seemed to divulge the best results. The complement of the yellow highlighted sequences seemed to be like Ori H 14, and the complement of the blue highlighted sequence near the anticodon region seemed to be similar to CSB-3/Ori L. In the diagrams below the figure, capitalized nucleotide letters were kept in tRNA and lowercase nucleotide letters are clipped out at the intron/exon junction.

The following grids were the results expanded from the above figures: It should be noted that the numbering is for the final tRNA only, and those sequences that were clipped out were not numbered in the figure used. The following two sequences were found approximately 70 bases apart.
Better results were found using RNA complements as well as DNA complements because they allowed for g-u as well as g-c bonds. Although this may or may not reflect actually combinations in the world of molecular biology, the following were found. Using these complements it is important to note that the complement of the pyrimidine tract used in intron/exon cleavage is a ggggg sequence.
Sulfolobus tokodaii pre-tRNA-Lys (UUU)
3’ |
70 |
|
|
|
|
|
|
|
|
|
|
#59 |
|
|
|
|
C |
G |
G |
G |
C |
G |
G |
C |
C |
C |
U |
A |
|
|
|
|
G |
C |
U |
U |
G |
C |
C |
G |
G |
G |
A |
T |
|
|
complement |
|
G |
C |
T |
T |
G |
C |
|
G |
G |
G |
A |
T |
|
|
Ori H H14 |
|
T |
|
|
|
|
|
G |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
c |
u |
u |
g |
c |
|
g |
g |
g |
a |
u |
g |
|
CSB-1 |
|
|
|
|
|
|
|
g |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5’ |
#32 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
C |
U |
U |
U |
U |
A |
a |
c |
c |
g |
c |
g |
u |
a |
|
|
|
G |
G |
G |
G |
U |
u |
g |
g |
u |
g |
u |
a |
|
complement |
|
|
G |
G |
G |
G |
T |
t |
g |
g |
t |
g |
t |
a |
|
CSB-3 |
|
|
G |
G |
G |
G |
T |
t |
g |
g |
t |
g |
t |
a |
|
Ori L UMS |
|
|
g |
g |
g |
g |
t |
t |
g |
g |
|
|
|
|
|
Telomere repeat |
S. tokodaii pre-tRNA-Ser (UGA)
3’ |
70 |
|
|
|
|
|
|
|
|
|
|
#59 |
|
|
|
|
C |
G |
G |
G |
C |
G |
G |
C |
C |
C |
U |
A |
|
|
|
|
G |
C |
U |
U |
G |
C |
C |
G |
G |
G |
A |
T |
|
|
complement |
|
G |
C |
T |
T |
G |
C |
|
G |
G |
G |
A |
T |
|
|
Ori H H14 |
|
T |
|
|
|
|
|
G |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
c |
u |
u |
g |
c |
|
g |
g |
g |
|
|
|
|
CSB-1 |
|
|
|
|
|
|
|
g |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
3’ |
#34 |
|
|
|
|
|
|
|
|
#25 |
|
|
|
|
|
|
U |
U |
C |
C |
G |
G |
U |
C |
G |
G |
G |
|
|
|
|
|
G |
G |
G |
G |
U |
U |
G |
G |
T |
T |
T |
|
|
|
complement |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
G |
G |
G |
G |
T |
T |
G |
G |
T |
|
T |
|
|
|
CSB-3/OriL |
|
|
|
|
|
|
|
|
|
|
G |
|
A |
|
|
|
|
G |
G |
G |
G |
T |
T |
G |
G |
|
|
|
|
|
|
Telomere repeat |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
S. tokodaii pre-tRNA-Thr (UGU)
3’ |
#70 |
|
|
|
|
|
|
|
|
|
|
#59 |
|
|
|
C |
G |
G |
C |
G |
A |
C |
G |
C |
C |
C |
C |
U |
A |
|
|
G |
G |
C |
G |
U |
U |
G |
C |
G |
G |
G |
G |
A |
T |
|
complement |
G |
T |
C |
|
T |
T |
G |
C |
G |
G |
G |
G |
A |
T |
|
Ori H H14 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
t |
c |
|
u |
u |
g |
c |
g |
g |
g |
g |
a |
u |
g |
CSB-1 |
|
|
|
|
|
a |
c |
g |
c |
c |
c |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
5’ |
#24 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
G |
C |
G |
C |
C |
G |
G |
C |
C |
U |
U |
|
|
|
|
|
U |
G |
U |
G |
G |
U |
U |
G |
G |
G |
G |
|
|
|
Complement |
|
T |
G |
T |
G |
G |
T |
T |
G |
G |
G |
G |
|
|
|
CSB-3/Ori L |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CSB-1 L. tarentolae sequence from Gao, G. et. al. (2001)
In the above tRNAs the CSB-1 like sequences were all in the acceptor arm and remained intact after the tRNA was processed. The CSB-3 complementary sequences encompassed an intron/exon junction in all cases and nucleotides were removed to generate the finished tRNA.
Other tRNAs were searched but the locations of complementary sequences were not as consistent as the pre-tRNA examples above:
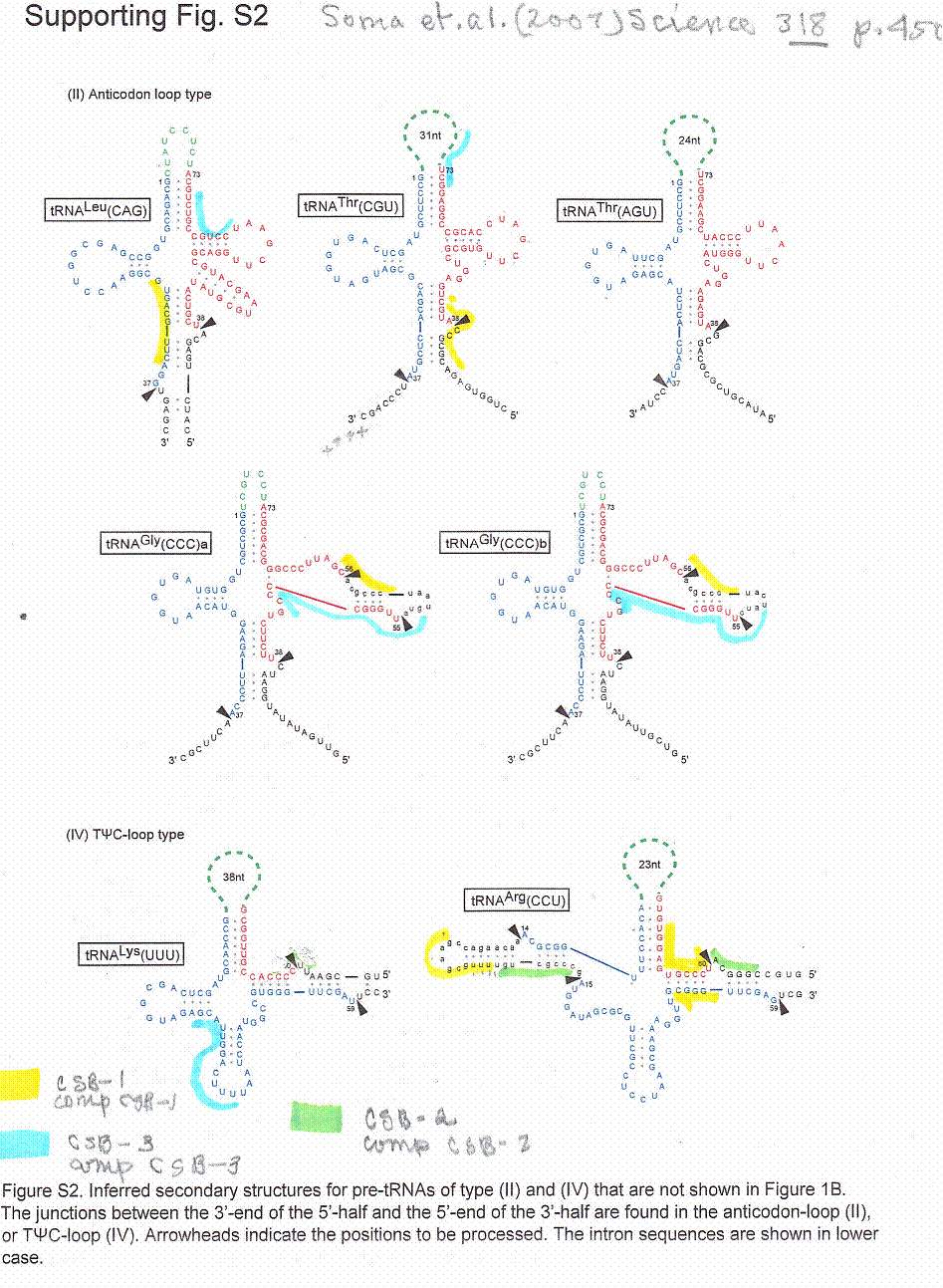
The putative pre-tRNA circles of C. merolae, a red alga, are grouped into four different types which are thought to be the result of linear sequences where the 3’ end of the molecule is actually 5’ to the 5’ end of the tRNA (Soma, A. et. al. (2007)). The linear sequence is then circularized and refashioned (to use a non-scientific word) into a tRNA. Below is a series of putative tRNAs with CSB-1 and CSB-3 like complement sequences highlighted by this author

In a search for CSB like sequences some similarities were found. (Sequences for the 74 and 31 nucleotide loops between #1 and #73 were from another figure)
tRNA gln (CUG)
A |
U |
U |
C |
C |
C |
G |
A |
U |
G |
G |
G |
U |
C |
C |
U |
3’ #73 |
|
t |
a |
g |
g |
g |
g |
c |
u |
g |
u |
u |
c |
a |
g |
g |
a |
Complem. |
|
t |
a |
g |
g |
g |
g |
c |
|
g |
t |
t |
c |
|
g |
|
|
Ori H 14 |
|
|
|
|
|
|
|
|
|
|
|
|
|
t |
|
|
|
|
|
|
|
|
c |
c |
c |
g |
|
c |
a |
|
|
|
|
|
|
|
|
|
|
g |
g |
g |
g |
c |
|
g |
t |
t |
c |
|
|
|
|
CSB-1 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
C. merolae pre-tRNA ala (UGC) had a cleavage point next to the 60th nucleotide. Uu (and preceding nucleotides 5’) are clipped out of the pre tRNA molecule as part of the processing which generates tRNA ala (UGC)
|
|
#60 |
|
|
|
|
|
|
|
|
|
|
|
#73 |
|
u |
u |
U |
C |
C |
C |
G |
C |
C |
G |
C |
C |
U |
C |
A |
3’ |
a |
a |
g |
g |
g |
g |
u |
g |
g |
u |
g |
g |
a |
g |
u |
RNA complement |
|
|
g |
g |
g |
g |
u |
g |
g |
u |
g |
|
a |
|
|
CSB-3 like seq |
|
|
|
|
|
|
u |
|
|
|
|
u |
|
|
|
|
a |
a |
g |
g |
g |
g |
t |
g |
g |
t |
g |
|
a |
|
t |
Kinet. Gap seq |
|
|
|
|
|
|
t |
|
|
|
|
t |
|
a |
|
|
The UGC anticodon loop had the following sequence also:
|
|
|
|
|
|
|
|
|
|
|
|
c |
u |
u |
u |
g |
c |
a |
a |
g |
#39 |
|
|
g |
g |
g |
g |
c |
g |
u |
u |
c |
|
|
RNA complement |
g |
g |
g |
g |
c |
g |
u |
u |
c |
|
|
CSB-1 |
|
c |
c |
c |
g |
c |
a |
|
|
|
|
Ori H hexamer |
|
|
|
|
|
|
|
|
|
|
|
|
tRNA leucine (UAA)
3’ |
|
|
|
|
|
|
|
|
|
#73 |
5’ |
|
c |
u |
u |
c |
a |
g |
u |
c |
a |
c |
A |
U |
|
g |
g |
g |
g |
t |
t |
g |
g |
t |
g |
t |
a |
RNA complement |
g |
g |
g |
g |
t |
t |
g |
g |
t |
g |
t |
a |
CSB-3 like |
|
|
|
|
|
|
|
|
|
|
|
|
|
tRNA leucine (UAA)
5’ |
|
|
|
|
|
|
|
|
|
|
#21 |
|
3’ |
|
|
u |
g |
c |
u |
c |
c |
a |
c |
g |
c |
A |
A |
G |
|
|
g |
t |
g |
g |
g |
g |
t |
g |
c |
g |
t |
t |
c |
RNA complement |
|
g |
t |
|
a |
g |
g |
|
g |
c |
g |
t |
t |
c |
Ori H H 14/CSB-1 |
|
|
|
|
|
|
|
g |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Etc.
Some CSB-like sequences encompassed junctions and others did not.
Other CSB like sequences were found in more C. merolae tRNAs
Of interest to note is that the complementary CSB like sequences for C. merolae were not found at the same location in each tRNA, unlike the archael tRNAs described above and below..
Fujishima, K. et. al. (2009), doing a closely detailed study of tRNA “fragments” in Archaea (which will not be described here) compared tRNA intron/exon junctions with “leader” junctions to show that they were alike and had similar sequences. For our purposes, the junction sequences for both the intron and leaders seem to be very like RNA complements of CSB-1 and CSB-2 and look to be an RNA palindrome. (cguuucggggcg) Please see below;
It does appear that CSB-like complementary sequences are located at the same locations near intron/exon junctions (29/30) in these archaea.

It does appear that CSB-like complementary sequences are located at the same locations near intron/exon junctions (29/30) in these archaea.
In a cluster of nine linked tRNA genes in B. subtilis the following were found at the 5’ end of the tRNA leu (TAA) DNA gene (Green, C and Vold, B. (1992):
From fig. 2
|
#584 |
|
|
|
|
|
|
|
|
|
|
|
#596 |
|
|
g |
g |
g |
g |
t |
g |
g |
t |
g |
g |
a |
a |
t |
tRNAleu(TAA) |
|
g |
g |
g |
g |
t |
g |
g |
t |
g |
|
a |
|
|
CSB-3 |
|
|
|
|
|
t |
|
|
|
|
t |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
g |
g |
g |
g |
t |
g |
g |
t |
g |
|
a |
a |
t |
Kinet gap seq (OriL) |
|
|
|
|
|
t |
|
|
|
|
t |
|
|
|
|
32 bases upstream was the sequence in the tRNA gly molecule
|
#546 |
|
|
|
|
|
#552 |
|
|
t |
c |
c |
c |
g |
t |
c |
tRNAGly (GCC) |
|
t |
c |
c |
c |
g |
t |
c |
CSB-2 |
|
|
|
|
|
|
t |
|
|
|
|
|
|
|
|
|
|
|
And, 8 nucleotides upstream from that, still in tRNAGly (GCC) was found a sequence that hinted of CSB-1, in reverse orientation:
|
#530 |
|
|
|
|
|
|
|
|
#539 |
|
|
g |
g |
t |
c |
g |
c |
g |
g |
g |
t |
tRNA gly (GCC) |
|
|
g |
t |
c |
g |
c |
g |
g |
g |
t |
CSB-1 T. brucei, T. equiperdum |
|
c |
|
|
|
|
|
|
|
|
|
|
|
|
|
a |
|
c |
g |
c |
c |
c |
|
Ori H hexamer |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
tRNA Primers and CSB-like sequences
So, it can be shown that CSB-like sequences are in tRNAs as well as kinetoplastid minicircles. Since CSB-3 and CSB-1 in kinetoplasts are primer sites for RNA primed DNA this author did a search for CSB-like sequences in tRNAs that are known to be primers for reverse transcription in retroviruses. A lot of work has been done to show that the RNA primer, lysine-3 tRNA, and to a lesser extent other tRNAs, complement retroviruses at a “Primer Binding Site” (PBS). A closer look at the PBS/tRNA lysine 3 site revealed the following (HIV-1 PBS and lys-3tRNA sequence from Beerens, N and Berkhout, B. (2002)) Additionally, tRNA lysine 3 is also attached to HIV-1 via a PAS site
|
Lys 3 tRNA |
c |
c |
a |
g |
g |
g |
t |
Ψ |
c |
a |
a |
g |
u |
c |
c |
c |
u |
g |
u |
u |
c |
g |
g |
g |
c |
g |
c |
c |
a |
3’ |
|
|
|
HIV-1 PBS |
|
|
|
|
|
|
|
|
|
|
|
c |
a |
g |
g |
g |
a |
c |
a |
a |
g |
c |
c |
c |
g |
c |
g |
g |
u |
g |
a |
c |
|
RNA comp |
|
|
|
|
|
|
|
|
|
|
|
g |
u |
c |
c |
c |
u |
g |
u |
u |
c/u |
g |
g |
g |
c |
g |
u |
u |
g |
c |
u |
g |
|
CSB-1 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
t |
g |
g |
g |
c |
g |
t |
t |
|
c |
|
|
|
Ori H H14 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
g |
t |
|
|
g |
g |
g |
c |
g |
t |
t |
|
c |
t |
g |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
a |
g |
|
|
|
|
|
|
|
|
|
|
|
|
Ori H hexamer |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
c |
c |
c |
g |
c |
a |
|
|
|
|
|
|
CSB-2 C. fasc. |
|
|
|
|
|
|
|
|
|
|
|
|
t |
c |
c |
c |
|
g |
t |
t |
c |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Lys 3 tRNA anti PAS site |
c |
c |
a |
g |
g |
g |
t |
Ψ |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CSB-2 C. fasc. |
|
|
t |
c |
c |
c |
g |
|
|
t |
t |
c |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
The same PBS:lystRNA-3 complementation is used for reverse transcription in feline immunodeficiency virus (FIV), and equine infectious anemia virus (EIAV) genomic RNAs (Arts, E. J. et. al (1996))
So, now let’s take a look at some of the bcl translocations through the prism of an RNA complement
|
Genbank U73664 Multiple Myeloma |
||||||||||||||||||||||||||||
|
Bcl locus on 11.q13 |
Gamma switch region 14q32 |
|||||||||||||||||||||||||||
|
|
c |
t |
g |
t |
t |
c |
c |
c |
g |
c |
a |
g |
g |
t |
g |
a |
g |
c |
a |
g |
g |
g |
g |
c |
a |
g |
g |
t |
|
Part of PBS sequence |
c |
c |
c |
g |
c |
|
g |
g |
u |
g |
a |
|
c |
a |
|
|
|
|
|
|
|
|
|
|||||
|
CSB-2 T. congolese |
t |
c |
c |
c |
g |
|
a |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
t |
|
c |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
g |
g |
t |
a |
g |
g |
g |
g |
c |
g |
t |
t |
c |
g |
c |
u |
c |
g |
u |
c |
c |
c |
c |
g |
u |
u |
c |
g |
|
|
|
g |
t |
a |
g |
g |
g |
g |
c |
g |
t |
t |
c |
g |
|
Ori H H14/ CSB-1 |
|
|
|
|
||||||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
t |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Ori H Hexamer |
|
c |
c |
c |
g |
c |
a |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CSB-2 T. spp. |
|
c |
c |
c |
c |
g |
t |
t |
c |
|
||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
It is important to note that the above translocation created the OriH H14/CSB-1 sequence; a primer site.
A statistical analysis to show that these sequences are inherently significant has not been done. They are however conserved or selected for in some way because in both kinetoplasts and tRNAs they are found over and over again.
Of note it has also been reported that there is a conserved region in the chloroplast minicircles of a symbiotic dinoflagellate in coral where this author found sequences very similar CSB-1, CSB-2 and CSB 3 .(Moore. R. B. (2003) et. al. ) The minicircle contained one gene; psbA, but this author could not confirm the conserved sequences in any other papers describing dinoflagellate chloroplast minicircles, including GenBank submissions.
HIV-2 stem loop 1 packaging Signal
In a paper by Baig et. al. the authors study the sequence for what they call 5’pal and 3’pal; parts of the packaging signal. The complement of these two sequences together is CSB-1 and the other side of the helix is CSB-2’s complement. CCAUGG at the top loop is the kissing part of the kissing loop used to link two copies of single stranded RNA before encapsidation.

7S RNA
7S RNA also has CSB-1, CSB-2 and CSB-3-like sequences primarily in its first 40 nucleotides (sequence from Li, WY et. al. (1982)
5’ pppGCCGGGCGGU GCGCACGCCU GUAGUCCCAG CUACUCGGGA GGCU
CGGCCCGCCG CGCGUGCGGG UGUCGGGGUU GGUGAGCCCG UU complement
The authors of this early paper present a conserved homologous region to Alu (human) RNA, B1 family (?) RNA and La 4.5S RNA. (NOT what is considered the ALU domain of SRP) Of note is the comparison between Alu and 7s because, although not a wonderful fit, it is reminiscent of PBS and the inverted repeats at the 29/30 junction of archael pre-tRNA described by Fujishima above. The bolded nucleotides are conserved and encompassed between two Okazaki fragments.
|
7S RNA (rat) |
250 |
C |
C |
A |
C |
U |
G |
C |
A |
C |
U |
C |
C |
A |
G |
C |
C |
U |
G |
G |
G |
C |
A |
A |
C |
A |
U |
A |
G |
C |
G |
|
|
B1 mOUSE |
|
|
|
|
|
|
|
|
|
|
|
C |
C |
A |
G |
C |
C |
U |
G |
G |
|
|
|
|
|
|
|
|
|
|
|
|
|
La 4.5S RNA |
|
|
|
|
|
|
|
|
|
|
|
C |
C |
A |
G |
C |
C |
U |
G |
G |
G |
C |
U |
A |
C |
A |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Alu (human) |
241 |
C |
C |
A |
C |
U |
G |
C |
A |
C |
U |
C |
C |
A |
G |
C |
C |
U |
G |
G |
G |
U |
G |
A |
C |
A |
G |
A |
A |
C |
A |
|
|
COMPLEMENT |
|
|
|
|
|
G |
U |
G |
U |
C |
G |
G |
G |
U |
C |
G |
G |
G |
C |
C |
C |
G |
C |
U |
G |
U |
U |
|
|
|
|
|
|
PBS |
|
|
|
|
|
g |
u |
|
u |
c |
g |
g |
g |
|
c |
|
|
g |
c |
c |
c |
g |
c |
|
g |
u |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
a |
|
|
|
a |
|
a |
a |
|
|
|
|
|
|
g |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
While these sequences are similar to an alu sequence, they are not considered the Alu domain that occurs at the 5’ end of the SRP molecule. (Huck et. al. (2004) where CSB-1,2, and 3 similar sequences were found by this author. A truncated SRP molecule where only the Alu domain remains, binds to heterodimeric SRP 9/14 at the bolded nucleotides GCC (Weichenrieder, O. et. al. (2001)) and at UC in the 3’ domain (not shown) and are thought to be involved with temporarily halting translation while the protein complex is lined up with a membrane pore.
|
Helix 2 and u turn of 5’ ALU domain –binding site of SRP9/14 |
|||||||||||||||||
|
#13 |
g |
g |
c |
g |
c |
g |
c |
g |
c |
c |
u |
g |
u |
a |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
RNA comp |
t |
t |
g |
c |
g |
t |
g |
c |
g |
g |
g |
t |
a |
t |
|
|
|
|
CSB-1 T. equiperdum |
|
|
|
c |
g |
t |
g |
c |
g |
g |
g |
t |
a |
|
|
|
|
CRSPRS
CRSPRS are inverted repeats found in Bacteria and Archaea that provide “immunity’ by capturing bits of nucleic acids from “invading” plasmids and viruses. These nucleic acids are then replicated and passed down to the next generation as a memory of previous exposures. Without going into detail, the authors of a recent paper, Horvath, P and Barrangou, R. (2010)) found four sets of CRSPRS (clustered interspersed short palindromic repeats) in a strain of Streptococcus thermophilus. The stem loop configuration of each set of CRSPRS
|
T |
G |
T |
A |
C |
T |
C |
T |
C |
A |
A |
G |
A |
T |
T |
T |
|
CRSPR 1 |
|||||
|
G |
T |
G |
T |
G |
G |
G |
G |
G |
T |
T |
C |
T |
G |
G |
G |
|
RNA complement |
|||||
|
|
|
|
|
|
|
|
|
|
|
|
C |
U |
G |
G |
G |
|
Primer site |
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
G |
T |
A |
|
G |
G |
G |
|
G |
T |
T |
C |
T |
G |
|
|
|
CSB-1/Ori H-14 |
|||||
|
|
|
|
G |
|
|
|
C |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Inverted rpt |
Acquired nucleotides |
|
|
|
|
|
|
|
||||||||||||||
|
T |
T |
A |
C |
C |
T |
C |
G |
A |
G |
A |
R |
G |
G |
G |
A |
C |
G |
G |
A |
A |
A |
C |
|
CRSPR 2 |
|||||
|
G |
G |
T |
G |
G |
G |
G |
T |
T |
T |
T |
T |
C |
T |
T |
T |
G |
C |
C |
T |
T |
T |
G |
|
RNA complement |
|||||
|
|
G |
T |
G |
G |
G |
G |
C |
|
|
T |
T |
C |
T |
|
|
|
|
|
|
|
|
|
|
CSB-1/Ori H H-14 |
|||||
|
|
|
A |
|
|
|
|
|
|
G |
|
|
|
|
G |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
C |
T |
T |
G |
C |
C |
|
T |
|
|
|
CSB-2 T. brucei |
|||||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
C |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Inverted rpt |
Acq. Nuc. |
Inverted rpt |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|||||||||
|
G |
C |
T |
G |
T |
G |
T |
T |
G |
T |
T |
T |
C |
|
|
CRSPR 3 |
|||||||
|
|
C |
T |
G |
T |
G |
|
|
|
|
|
|
|
|
|
Primer site |
|||||||
|
C |
G |
A |
T |
G |
T |
G |
G |
T |
G |
G |
G |
G |
|
|
RNA complement |
|||||||
|
|
|
A |
T |
G |
T |
G |
G |
T |
G |
G |
G |
G |
|
|
Csb-3 |
|||||||
|
|
|
|
|
|
|
|
|
T |
|
|
|
|
|
|
|
|||||||
|
t |
|
a |
t |
g |
t |
g |
g |
t |
g |
g |
g |
g |
|
|
Kinetoplastid gap |
|||||||
|
|
a |
|
|
|
|
|
|
t |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Inverted repeat |
Acquired nucleotides |
|
|
|
|
|
|
|
|
|
|
|||||||||||
|
T |
C |
C |
C |
G |
C |
A |
C |
A |
C |
G |
C |
G |
G |
G |
G |
G |
|
|
CRSPR 4 |
|||||
|
G |
G |
G |
G |
C |
G |
T |
G |
T |
G |
C |
G |
C |
C |
C |
C |
C |
|
|
RNA complement |
|||||
|
G |
G |
G |
G |
C |
G |
T |
G |
|
|
|
|
|
|
|
|
|
|
|
CSB-1 |
|||||
|
|
|
|
|
|
|
|
|
C |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
G |
T |
G |
C |
C |
C |
|
|
|
|
CSB-2 |
|||||
|
|
|
|
|
|
|
|
|
C |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Inverted repeat |
Acq. Nuc |
Inverted repeat |
|
|
|
|
|
|
|
|
||||||||||||||
If one is willing to believe that the complement of these sequences are CSB-like, then it is of use to note that they are created by the actual recombination event that took place. CRSPRS are associated with CAS’s, gene families, that, taking an educated guess, are probably involved in defending the cell against re-invasion of alien molecules. More can be found in the article mentioned above.
Conclusions
Illegitimate translocations are usually thought to be the result of an incorrect VDJ recombination in leukemias and myelomas involving immunoglobulins. While this may be true, and proven scientifically in some cases, there are many translocations where one cannot find the hexamer and nonamer used as binding sites for VDJ recombinase, and that have extra bases (“N” sequences, “intervening sequences” etc.) that are present in the new, aberrant result.
At least for some bcl2 and bcl6 translocations there seems to be either the creation or involvement of sequences closely resembling the conserved sequences CSB-1, CSB-2 and CSB-3. CSB-1 and CSB-3 are found in kinetoplasts as RNA-primed origins of replication for H and L strands that use a replication mechanism similar to lagging strand synthesis. Looking at it another way, CSB-1 and CSB-3 are RNA primer sites, with CSB-3 having a special function during replication. While this says nothing about bcl translocations it is useful to find that after the recombination process, that sequences similar to Ori H H14 and Ori L (CSB-1 and CSB-3) are moved closer together.
The fact that these translocations occur at “low” rates in healthy patients shows that there is a constitutive mechanism functioning. The question is, is it mediated by VDJ recombinase? If not, what are its origins? Does it have something to do with primer initiation of replication?
CSB-1 and CSB-3 are both bound by UMSBP a protein involved in the replication of kinetoplastid DNA minicircles. Whether these conserved sequences existed in a world without DNA and proteins and were involved in replication remains to be shown, but they are definitely found in RNA molecules. (pre-tRNAs, tRNAs, 5’ end of cloverleaf of polio virus and other RNA enteroviruses SRPs etc.) Complements of the conserved sequences also are closely related to intron/exon excision and ligation and can be demonstrated in some archael pre-tRNAs.
The fact that these sequences are found as complements of some pre-tRNA molecules is a topic that is too broad to discuss here, but it has been postulated that the 3’ end of tRNA (acceptor and Tψ arms) is an ancient molecule that predates the anticodon and D arms needed for translation and was involved in replication.( Maizels, N. and Weiner, A. (1994)). This is probably too simplistic as tRNAs may be constructed from intron ligations at 2 or 3 or more sites. But it is useful to note that the 5’ cloverleaf secondary formation of polio RNA virus which is responsible for the switch from replication to (parasitic) translation contains a sequence similar to the complement of CSB-3 at the 5’ end.
Important is the fact that CSB-like sequences are found in tRNA primer binding sites used in the reverse transcription of retroviruses; another form of RNA primed replication.
The ability to see these sequences by using RNA complements hints at an RNA origin for the CSB sequences.
REFERENCES
Abu-Eineel, K et. al. (1999) Universal minicircle sequence-binding Protein, a Sequence-specific DNA-binding Protein that Recognizes the Two Replication Origins of the Kinetoplast DNA Minicircle JBC vol 274 p. 134-19-13426
Arts, E. J. et. al. (1996) Initiation of (-) strand DNA synthesis from tRNA lys3 on lentiviral RNAs: Implications of specific HIV-1 RNA-tRNA lys 3 interactions inhibiting primer utilization by retroviral reverse transcriptases, PNAS, vol 93, pp 10063-10068.
Beerens, N. and Berkhout, B. (2002), The tRNA Primer Activation Signal in the Human Immunodeficiency Virus Type 1 Genome Is Important for Initiation and Processive Elongation of Reverse Transcription, Journal of Virology, vol 76, No.5, p.2329-2339.
Campbell, T. G and Cech, T. R. (1995) Identification of ribozymes within a ribozyme library that efficiently cleave a long substrate RNA, RNA vol 7, 598-609.
Cleary, M. and Sklar, J, (1985) Nucleotide sequence of a t(14:18) chromosomal breakpoint in follicular lymphoma and demonstration of a breakpoint cluster region neat a transcriptionally active locus on chromosome 18. PNAS vol 82, pp.7439-7443
Frick, D. and Robinson, C., (2001), DNA Primases, Ann. Rev Biochemistry, vol 70, 39-80.
Gao, G. et. al., (2001), Guide RNAs of the recently isolated LEM 125 strain of Leishmania tarentolae: an unexpected complexity, RNA vol 7 p.1335-1347.
Green, C. and Vold, B (1992), A Cluster of Nine tRNA Genes between Ribosomal Gene Operons in Bacillus subtilis, Journal of Bacteriology, vol 174, No. 10, pp3147-3151
Gu, Y. et. al (1992) The (4:11) (q21:23) chromosome translocations in acute leukemias involve the VDJ recombinase, PNAS vol 89 pp10464
Hines, J.C. and Ray, D. S. (2008) Structure and Discontinuities of kinetoplast DNA associated minicircles during S. Phase in Crithidia fasciculata, NAR vol 36, 444-450.
Hines, J.C. et. al. (2001) RNA primer removal and gap filling in a model minicircle replication intermediate, Mol. Biochem Parasitology vol 115, p. 63-67
Horvath, P. and Barrangou, R., (2010), CRISPR/cas, the Immune System of Bacteria and Archaea, Science, vol 327, p. 167.
Huck, et. al. (2004), Conserved tertiary base pairing ensures proper RNA folding and efficient assembly of the signal recognition particle Alu domain, Nucleic Acids Research vol 32(16), pp. 4915-4924
Jasmer, D.P. and Stuart, K. (1986), Mol. Bioch Parasitology, vol 18, no. 3, pp. 257-269
Ji, W. et. al. (1995) Frequent Detection of bcl2/JH Translocations in Human Blood and Organ Samples by a Quantitative Polymerase Chain Reaction Assay, Cancer Research vol 55, 2876-2882.
Lukes, Julius (1998), Pankinetoplast DNA structure in a primitive bodonid flagellate Cryptobia helicis, The Embo Journal vol 17, 838-846.
Marck, C. and Grosjean, H., (2003), Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications, RNA, vol 9, 1516-1531.
Maizels, N. and Weiner, A. (1994), Phylogeny from function: Evidence from he molecular fossil record that tRNA originated in replication, not translation, PNAS vol 91, pp. 6729-6733
Li, Wen-Yu et. al. (1982), Nucleotide Sequence of 7SRNA, Homology to ALU DNA and LA 4.5 S RNA, JBC vol 257, pp 5136-5142.
Moore, R.B. et. al. (2003) Highly organized structure in the non-coding region of the psbA minicircle from clade C Symbodium, International Journal of Systematic and Evolutionary Microbiology, vol 53, 1725-1734.
Onn, I. et. al., (2006), Binding of the Universal Sequence Binding Protein at the kinetoplast DNA Replication Origin, The Journal of Biological Chemistry, vol 281, pp, 37468-37476.
Ntambi, J and Englund, Paul T. (1985), A Gap at a Unique Location in Newly Replicated Kinetoplast DNA Minicircles from Trypanosoma equiperdum, The Journal of Biological Chemistry, Vol 260. pp5574-5579
Qimron, U. et. al., (2006) Primer Initiation and Extension by T7 DNA primase, the EMBO Journal vol 25, 2199-2208
Ray, Dan S., (1989), Conserved Sequence Blocks in Kinetoplast Minicircles from Diverse Species of Trypanosomes, Molecular and Cellular Biology, vol 9, 1365-1367.
Shapiro, T. and Englund, PT (1995), The Structure and Replication of Kinetoplast DNA, Annual Review of Microbiology, vol 49, 117-43
Soma, A., et. al. (2007) , Permuted tRNA Genes Expressed via a Circular RNA Intermediate in Cyanidioschyzon merolae, Science, vol 318, p. 450-453.
Tzafti et. al., (1995) Universal Minicircle Sequence Binding Protein, a CCHC-Zinc finger protein that binds the Universal Minicircle Sequence of Trypanosomatids, vol 270, pp. 21339-21345.
Weichenrieder, O. et. al. (2001) Assembly of the Alu domain of the mammalian signal recognition particle, RNA, vol 7, pp731-740.
Ye, B. H. et. al. (1995) Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B-cell lymphoma EMBO J. vol 14 6209-6217